The reconstruction of organism-specific metabolic networks is at the heart of systems biology. GLOBUS is a method for the probabilistic, genome-wide annotation of metabolic genes. All genes displaying sequence similarity to known enzymes are considered simultaneously. This makes it possible to calculate the probability for the association of every gene with a set of candidate functions given that all other genes can also be assigned to one or more possible activities. The method combines sequence similarity with gene-gene functional associations, these include phylogenetic correlations, chromosome gene clustering, and gene co-expression. These network context correlations provide crucial evidence to decide the correct function for genes with low sequence similarity to known enzymes.
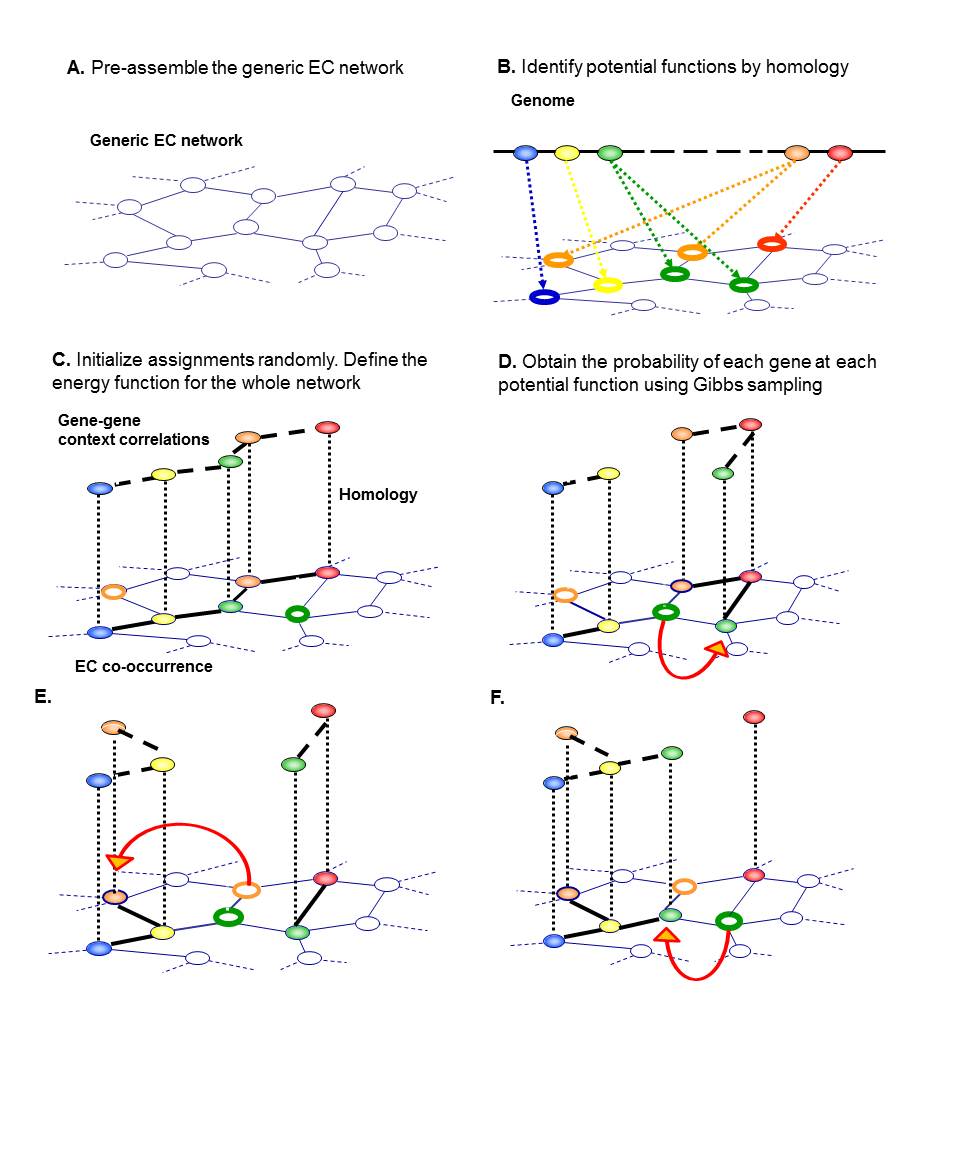
The conceptual outline of GLOBUS is shown in the figure below. First, we assemble a generic reaction network containing all metabolic activities characterized in the Enzyme Commission (EC) system (a). Based on sequence homology to well annotated enzymes, possible functions are identified for each gene (b); an initial network assignment is made by randomly selecting one such function for each gene. For every assignment of genes to functions in the generic reaction network we define a score; this score is higher if genes have good context correlations with their neighbors and high homology to their assigned locations (c). Based on this joint score, we use Gibbs sampling to derive the marginal probabilities of each gene at each of its candidate functions (d-f). As shown in the figure we pick genes one at a time; we then re-assign each gene to one of its candidate functions depending on the global score at each of these positions given the current location of all other genes. At each step, a gene has a higher probability to be assigned to the function where it has a higher combined score of sequence homology and context correlations. This procedure is repeated for a number if iterations until convergence to the desired marginal probabilities.
|
