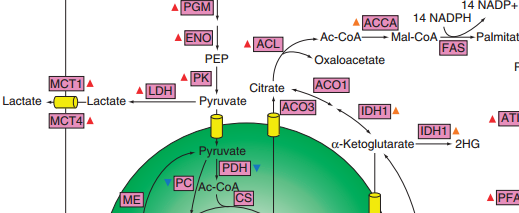
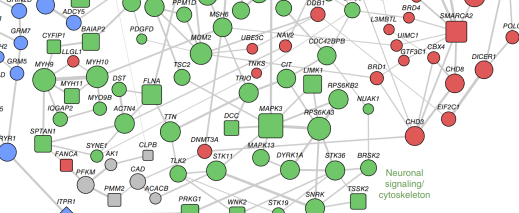
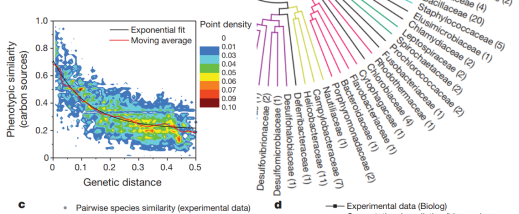
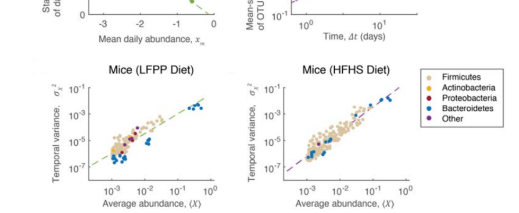
Dennis Vitkup, PhD
Associate Professor Department of Systems Biology / Department of Biomedical Informatics Columbia University 1130 St. Nicolas Avenue, New York, NY 10032 Phone: 212-851-5151 - Fax: 212-851-5149 E-mail: dv2121 [at] cumc [dot] columbia [dot] eduOur systems biology laboratory is working on several important biological and biomedical questions. To address scientific challenges, we develop and apply novel computational, theoretical and experimental tools and approaches. We also work closely with experimental collaborators across the investigated research themes. In our research, we seek to gain mechanistic insights into fundamental biological phenomena and to understand specific biological mechanisms underlying human disorders.

Links:
Columbia University in the City of New York
Department of Systems Biology
Department of Biomedical Informatics