Research
I. Evolutionary Systems Biology.
A rapidly increasing ability to sequence and edit genomes, to measure and perturb molecular and cellular phenotypes, and to computationally analyze these data, have significantly influenced evolutionary biology. The emerging fields of evolutionary systems and cell biology aim at understanding how the systems and physiological properties of cells and organisms affect evolutionary processes. In this research theme, we are interested in understanding how various levels of biological organization influence molecular and phenotypic evolution. . We investigate the interdependence between processes and constraints at various levels of biological organization. Our main hypothesis is that processes and interactions across biological scales play a major role in shaping the evolution of proteins, cellular networks, and ultimately species phenotypes. To invert the logic of the famous statement by Theodosius Dobzhansky, we believe that very little in protein and genomic evolution makes sense except in the light of the underlying biology.
II. Systems Biology of Neurodevelopmental and Psychiatric Disorders.
Over the last decade, amazing progress has been made towards understanding the biology of psychiatric and neurodevelopmental disorders. Recent studies by our group and colleagues demonstrated that common diseases of the mind are in fact brain diseases; i.e. they are often mediated by malfunctions of specific genes and biological processes, with contributions from various environmental factors. 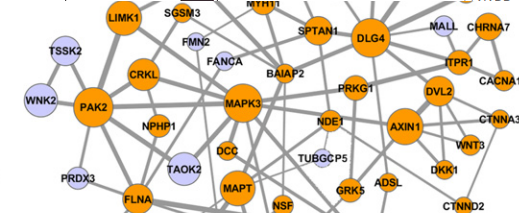 Our laboratory pioneered the development and application of several interrelated computational approaches that revealed key cellular networks associated with psychiatric disorders, such as autism and schizophrenia. We are currently interested in understanding specific cellular processes, cell types, and functional brain circuits that are perturbed in psychiatric disorders. We are also exploring how various genetic and environmental insults influence patients' phenotypes. Beyond understanding the genetics and systems biology of neurodevelopmental disorders, we believe that this research theme will shed light on normal brain operations, and reveal how the human brain integrates information and makes context-dependent decisions.
Our laboratory pioneered the development and application of several interrelated computational approaches that revealed key cellular networks associated with psychiatric disorders, such as autism and schizophrenia. We are currently interested in understanding specific cellular processes, cell types, and functional brain circuits that are perturbed in psychiatric disorders. We are also exploring how various genetic and environmental insults influence patients' phenotypes. Beyond understanding the genetics and systems biology of neurodevelopmental disorders, we believe that this research theme will shed light on normal brain operations, and reveal how the human brain integrates information and makes context-dependent decisions.
III. Cancer Cell and Tumor Metabolism.
To support the synthesis of biomass components and to generate the energy required to support proliferation, cancer cells significantly reshape functional properties and regulation of their metabolic networks. In this theme of our research we are investigating how tumors hijack normal cell metabolism to fuel cell survival and proliferation. 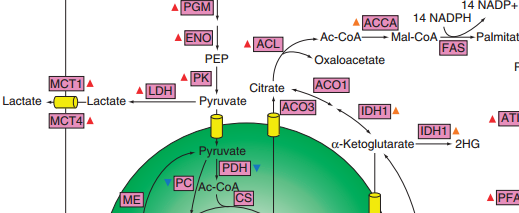 We build realistic biophysical models of cancer cells and simulate global metabolic flux distribution across tumors. We are also interested in understanding key regulatory and signaling pathways, such as PI3K/Akt and mTOR, underlying metabolic transformation in cancer cells. We specifically investigate what are the major metabolic bottlenecks for cancer cell growth across tumor types and environmental conditions. Our man hypothesis is that the availability of key nutrients, environmental conditions, and tissue microenvironment play major roles in determining metabolic flux distributions and proliferation properties of cancer cells. We also investigate how whole-body metabolism and different diets may influence cellular metabolism across tumor types.
We build realistic biophysical models of cancer cells and simulate global metabolic flux distribution across tumors. We are also interested in understanding key regulatory and signaling pathways, such as PI3K/Akt and mTOR, underlying metabolic transformation in cancer cells. We specifically investigate what are the major metabolic bottlenecks for cancer cell growth across tumor types and environmental conditions. Our man hypothesis is that the availability of key nutrients, environmental conditions, and tissue microenvironment play major roles in determining metabolic flux distributions and proliferation properties of cancer cells. We also investigate how whole-body metabolism and different diets may influence cellular metabolism across tumor types.
IV. Macroecological Dynamics and Interactions in Microbial Communities.
Bacterial species usually function in complex and dynamic communities (microbiomes). Microbes are also involved in multimodal interactions with each other, with the host, and with their environment. It is also now recognized that instability and dysbiosis of physiologically important communities may contribute to a wide range of human diseases, from immune disorders to psychiatric phenotypes to obesity. In this theme of our research we are interested in understanding the relationships and natural laws that describe the dynamics of microbial communities across space and time. We are also investigating metabolic interactions between bacteria in microbial communities. Our preliminary analyses revealed that the dynamics of microbiomes can be described by several robust statistical relationships that are strikingly similar to patterns previously observed across multiple other ecological systems. Using the computational reconstruction and simulation of microbial metabolism, we are investigating how functional properties of individual bacteria shape community-level ecological dynamics and synergistic interactions between microbiomes and host species.